Simulating Alanine dipeptide#
!pip install -q condacolab
import condacolab
condacolab.install()
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/condacolab.py:26
25 try:
---> 26 import google.colab
27 except ImportError:
ModuleNotFoundError: No module named 'google'
During handling of the above exception, another exception occurred:
RuntimeError Traceback (most recent call last)
Cell In[1], line 2
1 get_ipython().system('pip install -q condacolab')
----> 2 import condacolab
3 condacolab.install()
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/condacolab.py:28
26 import google.colab
27 except ImportError:
---> 28 raise RuntimeError("This module must ONLY run as part of a Colab notebook!")
31 __version__ = "0.1.6"
32 __author__ = "Jaime Rodríguez-Guerra <jaimergp@users.noreply.github.com>"
RuntimeError: This module must ONLY run as part of a Colab notebook!
%%capture
!conda install -c conda-forge openmm openmmtools mdtraj nglview
import openmm
from openmm import *
from openmm.app import *
from openmm.unit import *
import matplotlib.pyplot as plt
import mdtraj as md
import numpy as np
import pandas as pd
import openmmtools
from openmmtools import testsystems
Create alanine dipeptide all-atom simulation#
# Grab alaine dipeptide
dialanine = testsystems.AlanineDipeptideExplicit()
system, positions, topology = dialanine.system, dialanine.positions, dialanine.topology
from openmm.app import PDBFile
with open("dialanine.pdb", "w") as f:
PDBFile.writeFile(dialanine.topology, dialanine.positions, f)
#import nglview as nv
#pdb = md.load_pdb("dialanine.pdb")
#view = nv.show_mdtraj(pdb)
#view.add_ball_and_stick('all')
#view.center_view(zoom=True)
#view
Setup simulation in OpenMM#
# Simulation parameters for explicit solvent all-atom system
nonbondedMethod = PME
nonbondedCutoff = 1.0*nanometers
ewaldErrorTolerance = 0.0005
constraints = HBonds
rigidWater = True
constraintTolerance = 1e-5
hydrogenMass = 1.5*amu
dt = 2.0 * femtoseconds
temperature = 300*kelvin
friction = 1.0/picosecond
pressure = 1.0*atmospheres
barostatInterval = 25
equilibrationSteps = 10000
nsteps = 2500000 # ~5 ns if dt = 2 fs
#Reporters
dcdReporter = DCDReporter('trajectory.dcd', 1000)
dataReporter = StateDataReporter('log.txt', 1000,
totalSteps=steps,
step=True,
speed=True, progress=True,
potentialEnergy=True,
temperature=True,
separator='\t')
Run NPT simulation#
# Prepare the Simulation
system.addForce(MonteCarloBarostat(pressure, temperature, barostatInterval))
integrator = LangevinMiddleIntegrator(temperature, friction, dt)
integrator.setConstraintTolerance(constraintTolerance)
simulation = Simulation(topology, system, integrator)
simulation.context.setPositions(positions)
# Minimize
simulation.minimizeEnergy()
simulation.context.setVelocitiesToTemperature(temperature)
simulation.step(equilibrationSteps)
# Simulate
simulation.reporters.append(dcdReporter)
simulation.reporters.append(dataReporter)
simulation.step(nsteps)
Metadynamics simulation of alanine dipeptide#
# CV setup
num_grid_pts = 25
resolution = 2 * np.pi / num_grid_pts # bin width in radians
bias_factor = 3
# Torsion atom indices
psi_indices = [6, 8, 14, 16]
phi_indices = [4, 6, 8, 14]
# Define collective variables (CVs) as torsion angles
cv1 = CustomTorsionForce('theta')
cv1.addTorsion(*psi_indices)
system.addForce(cv1)
psi = BiasVariable(cv1, -np.pi, np.pi, resolution, True)
cv2 = CustomTorsionForce('theta')
cv2.addTorsion(*phi_indices)
system.addForce(cv2)
phi = BiasVariable(cv2, -np.pi, np.pi, resolution, True)
# Add barostat
system.addForce(MonteCarloBarostat(pressure, temperature, barostatInterval))
# Integrator and Simulation object
integrator = LangevinMiddleIntegrator(temperature, friction, dt)
integrator.setConstraintTolerance(constraintTolerance)
simulation = Simulation(topology, system, integrator)
simulation.context.setPositions(positions)
# Energy minimization and equilibration
simulation.minimizeEnergy()
simulation.context.setVelocitiesToTemperature(temperature)
simulation.step(equilibrationSteps)
# Set up Metadynamics
meta = Metadynamics(system,
[phi, psi],
temperature,
bias_factor,
height=1.0 * kilojoules_per_mole,
frequency=500, # bias added every 1 ps
save_frequency=5000, # optional: save bias periodically
bias_dir='bias_output')
# Reporters
simulation.reporters.append(DCDReporter('trajectory.dcd', 1000))
simulation.reporters.append(StateDataReporter(stdout, 1000,
step=True, potentialEnergy=True, temperature=True, progress=True,
remainingTime=True, speed=True, totalSteps=productionSteps, separator='\t'))
simulation.reporters.append(CheckpointReporter('checkpoint.chk', 5000))
# Run Metadynamics
meta.step(simulation, productionSteps)
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
/var/folders/3h/w4n54dz57dvbhgvz68gdv0f0f3hkmm/T/ipykernel_64866/2700539307.py in <cell line: 43>()
41 simulation.minimizeEnergy()
42 simulation.context.setVelocitiesToTemperature(temperature)
---> 43 simulation.step(equilibrationSteps)
44
45 # Set up Metadynamics
/opt/homebrew/anaconda3/envs/llpsmd3/lib/python3.9/site-packages/openmm/app/simulation.py in step(self, steps)
139 def step(self, steps):
140 """Advance the simulation by integrating a specified number of time steps."""
--> 141 self._simulate(endStep=self.currentStep+steps)
142
143 def runForClockTime(self, time, checkpointFile=None, stateFile=None, checkpointInterval=None):
/opt/homebrew/anaconda3/envs/llpsmd3/lib/python3.9/site-packages/openmm/app/simulation.py in _simulate(self, endStep, endTime)
204 stepsToGo = nextSteps
205 while stepsToGo > 10:
--> 206 self.integrator.step(10) # Only take 10 steps at a time, to give Python more chances to respond to a control-c.
207 stepsToGo -= 10
208 if endTime is not None and datetime.now() >= endTime:
/opt/homebrew/anaconda3/envs/llpsmd3/lib/python3.9/site-packages/openmm/openmm.py in step(self, steps)
11900 the number of time steps to take
11901 """
> 11902 return _openmm.LangevinMiddleIntegrator_step(self, steps)
11903
11904 def __init__(self, *args):
KeyboardInterrupt:
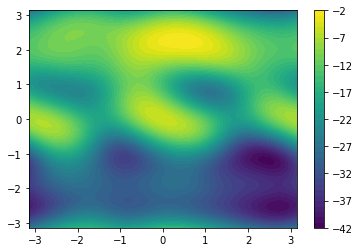
Analysis#
fes = meta.getFreeEnergy()
np.save('fes.npy', fes)
Z = np.load('fes.npy')
X, Y = np.linspace(-np.pi, np.pi, len(Z)), np.linspace(-np.pi, np.pi, len(Z))
X, Y = np.meshgrid(X,Y)
plt.contourf(X, Y, Z, 40)
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x7fe190c1f3d0>